COVID-19 research is performed in labs — but not the kind you might think. Thanks to the biomedical research platform Folding@home (F@h), computer labs operating out of regular people’s homes are actually running the complex calculations that will help scientists understand the coronavirus.
Yes, your friendly neighborhood laptop is capable of simulating COVID-19 protein dynamics. The goal, of course, is to understand how the virus behaves enough to develop the drugs that can stop it.
That’s why I’m thrilled to head up ServerCentral Turing Group’s effort to contribute to F@h’s research.
What is Folding@home, exactly?
Folding@home was a platform originally introduced by Stanford University and is now being maintained at Washington University. It is designed to provide a distributed supercomputer of sorts aimed at biomedical research. It normally focuses on diseases such as Parkinson’s and Alzheimer’s, but right now they working on COVID-19. Makes sense.
By recruiting an army of computers to work together, Folding@home creates a Franken-machine that’s more powerful than the biggest supercomputers in the world. (The more nodes, the better. And F@h is swimming in nodes.)
How does it work?
Anyone can download a piece of software on their computer (this can be a server, a desktop, a laptop, a game console…VMware has also released an OVA to install F@h on hypervisors and vSphere clusters). F@h also has support for discrete graphics cards, which are most commonly used for PC video games. Modern graphics cards are incredibly powerful and great at crunching numbers.
If there are “work units” available, your computer will download those work units, process them, and ultimately upload them back to F@h’s servers. Each “work unit” you complete gives you a certain number of points based on the difficulty. Those points are then credited to you or your team (if you’re a part of one).
Sounds great, what’s SCTG doing to contribute?
We’re taking several Dell R910s, as well as some R620 and R630 servers, and installing Linux on them with the F@h client. Each R910 has 40 processing cores. The R620s and R630s have somewhere between 12 and 28 cores each. We’ll add more as we can.
How do I check the stats?
There are two ways to see where we’re at:
- Go to the F@h website: https://stats.foldingathome.org/team/256378.
- Go to the Extreme Overclocking website (teams ranking in the top 12,000 show up here with advanced metrics): https://folding.extremeoverclocking.com/team_summary.php?s=&t=256378.
How do I get involved with Folding@home, too?
- Start out by creating a passkey at https://apps.foldingathome.org/getpasskey. This will make sure that points are credited to you as you finish work units.
- Pick a username (mine, for example, is Paul_Armenakis) and give it an email address (don’t worry, they don’t spam you). It takes a little while to get your passkey, so continue on to the next step while you wait.
- Go to https://foldingathome.org/alternative-downloads/ and pick your OS. It will download the installer, then install the program. After that is complete, it should launch a new web page, client.foldingathome.org, which is your locally installed client. If it just keeps refreshing, try a different browser.
- Be sure to change your identity from “Anonymous” to the username you already picked. Click Change Identity and enter in your Username, Team (if applicable), and Passkey (the passkey is important).
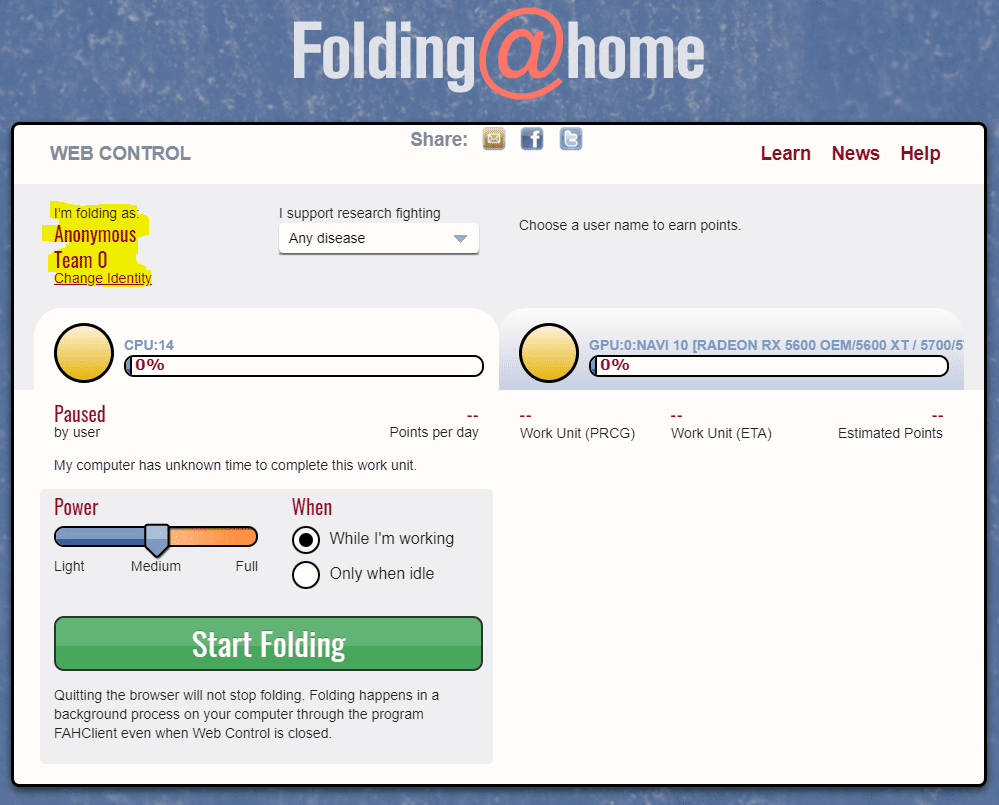
How do I change power usage settings?
You can change your settings for power usage and whether to wait until your computer is idle to fold. You can also set up remote management if you have multiple machines folding.
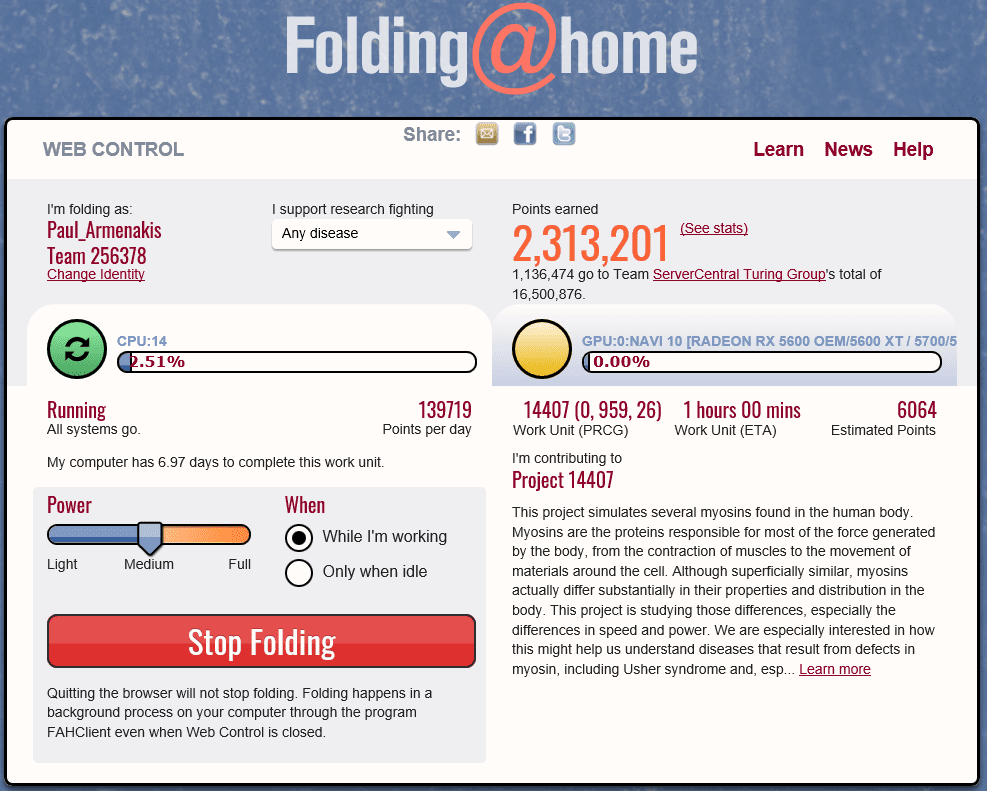
If you’re more of a power user, there’s also the Advanced Control, or FAHControl program, that you can use to control your environment. In Windows, right click the Folding@home icon on your taskbar and choose Advanced Control. This lets you set advanced settings on your local machine. And if you have multiple computers available to you, you can centrally control them all from this view.
Stay healthy, and happy folding!
